Paramyxoviridae
Paramyxoviridae (from Greek para- “by the side of” and myxa “mucus”) is a family of negative-strand RNA viruses in the order Mononegavirales.[1][2] Vertebrates serve as natural hosts.[3] Diseases associated with this family include measles, mumps, and respiratory tract infections.[4] The family has four subfamilies, 17 genera, and 78 species, three genera of which are unassigned to a subfamily.[5]
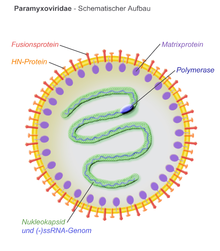
Virions are enveloped and can be spherical or pleomorphic and capable of producing filamentous virions. The diameter is around 150 nm. Genomes are linear, around 15kb in length.[6][1] Fusion proteins and attachment proteins appear as spikes on the virion surface. Matrix proteins inside the envelope stabilise virus structure. The nucleocapsid core is composed of the genomic RNA, nucleocapsid proteins, phosphoproteins and polymerase proteins.
The genome is nonsegmented, negative-sense RNA, 15–19 kilobases in length, and contains six to 10 genes. Extracistronic (noncoding) regions include:
Each gene contains transcription start/stop signals at the beginning and end, which are transcribed as part of the gene.
Gene sequence within the genome is conserved across the family due to a phenomenon known as transcriptional polarity (see Mononegavirales) in which genes closest to the 3’ end of the genome are transcribed in greater abundance than those towards the 5’ end. This is a result of structure of the genome. After each gene is transcribed, the RNA-dependent RNA polymerase pauses to release the new mRNA when it encounters an intergenic sequence. When the RNA polymerase is paused, a chance exists that it will dissociate from the RNA genome. If it dissociates, it must re-enter the genome at the leader sequence, rather than continuing to transcribe the length of the genome. The result is that the further downstream genes are from the leader sequence, the less they will be transcribed by RNA polymerase.
Evidence for a single promoter model was verified when viruses were exposed to UV light. UV radiation can cause dimerization of RNA, which prevents transcription by RNA polymerase. If the viral genome follows a multiple promoter model, the level inhibition of transcription should correlate with the length of the RNA gene. However, the genome was best described by a single promoter model. When paramyxovirus genome was exposed to UV light, the level of inhibition of transcription was proportional to the distance from the leader sequence. That is, the further the gene is from the leader sequence, the greater the chance of RNA dimerization inhibiting RNA polymerase.